Warning : This document is still a draft
Ensemble Clustering.
Ensemble clustering aims to combine multiple clustering together to get a better consensus clustering. Here, I will present the main approaches, and how evaluation is performed. I will introduce some reflection about scoring and diversity.
Introduction
In the previous post, we introduced shortly clustering. Here, this is about ensemble clustering.
Table of Content
TODO when article is not a draft anymore
Ensemble Clustering
It is possible to create multiple clustering, using different algorithms, different parameters, or different data processing. We can decide to select “the best clustering” among all that have been generated. Or we can decide to mix them together.
The goal of ensemble learning is to combine multiple base clustering to create a consensus clustering.
In this approach, we can expect to take the best of all results. In other words, a single clustering has its imperfections.
We will take the following example all along, with 8 items clustered three times:
- the two first time with three clusters,
- the last time with two clusters.
One possible outcome is the following:
This is one possible option. For instance, the partitioning n°2 is also a good candidate.
Big Data Clustering: Partitioning
Inspired by Ensemble Classification
This is mainly inspired by ensemble classification. Random forest is a good example. In a forest, the base classification algorithm are trees. A tree make rough classification: if \(x_i > 4\) then go left, else go right. These straight boundaries are not expressive enough, and make difficult generalization. By combining multiple trees, the boundary become “smooth” with a varied shape.
TODO: Illustration with anymated .gif
Diversity 1
In supervised ensemble learning, the assumption is that weak learners are uncorrelated in their errors. In practice, this is not often fulfilled, but if it were the case, theoretically, we could get very high accuracy (with an infinite number of classifiers). For ensemble clustering, “errors” cannot be measured, as the ground-truth clustering doesn’t exist. There is no way to verify that clusterings are uncorrelated in their errors.
Clustering algorithms, as well as classification algorithms, are very likely to converge to the same final state if nothing is done.
In other words, we would get correlated outcomes.
For instance, if you look at KMeans++ (default initialization procedure in sklearn), the probability to end up with exactly the same cluster centers is very high. This optimization of the KMeans algorithm enables to get very fast convergence due to optimized initialization, to the detriment of diversity.
The usual strategies to improve classifier diversity (which can be reused by ensemble clustering) are:
- Sampling (feature and/or item)
- Data projection (using random matrices), pre-processing (e.g., normalization)
- Changing algorithms parameters
- Using different algorithms
In some way, these operation are perturbation. If identified clusters are “true clusters”, they must be stable and recoverable even with the addition of noise.
With ensemble clustering, we can additionally play on the number of clusters. While there is many ways to improve diversity, most experimental protocols play on the cluster number to modify the outcome.
Ensemble Clustering Families
In the litterature, it is not clear how many families there are. We can categorized algorithms based on the intermediate structure between base and consensus clustering. We have:
- Item-based: the intermediate representation in a \(n \times n\) matrix representing the co-clustering frequency of items,
- Cluster-based: the representation is a \(d \times d\) matrix representing the overlap / similarity between clusters,
- Hybrid: a \(n \times d\) binary matrix represents the clusters in which an item has been clustered.
We will detail a bit these families with some example of algorithms.
Item-Based
This family is one of the most intuitive. The first step is to create the co-association matrix \(A\), which represents how many times item \(i\) has been clustered with item \(j\). For \(k\) based clustering, a term of this matrix is computed as:
\[A_{i, j} = \frac{1}{k} \sum_{\ell=1}^k \delta(C^{\ell}(i), C^{\ell}(j))\]with \(C^{\ell}(i)\) is the ID of the cluster of \(i\) in partitioning \(\ell\), and where
\[\delta(C^{\ell}(i), C^{\ell}(j)) = \left\{ \begin{array}{ll} 1 & \mbox{ if } C^{\ell}(i) = C^{\ell}(j) \\ e 0 & \mbox{ otherwise} \end{array} \right.\]Then, the second step is to cluster this matrix, with the goal to identify “blocks” of highly related items. In the oldest papers, they cluster this matrix using hierarchical agglomerative clustering.
These algorithms are very intuitive, as we stay at the item level and forget about base clusters at the start. However, the main problem is the scalability: the computation of the matrix \(A\) is computationally expensive and requires a lot of memory (scaling in \(\mathcal{O}(n^2)\). Therefore, it cannot scale to very large dataset (It is difficult to process more than 10,000 items on a regular computer).
Illustration
Using the initial plot, we get the following matrix:
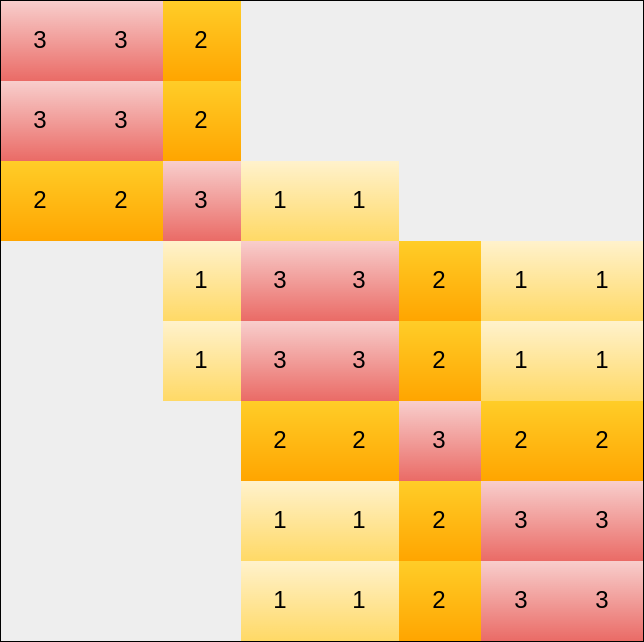
Note:
- Here, values are not normalized (you must divide all values by \(3\) to get values in \([0, 1]\)).
- Grey block represents \(0\)
Algorithms
- Evidence Accumulation Clustering (WEA Weighted evidence accumulation,)
Cluster-Based
Compare to the previous one, this family is very efficient in terms of scaling.
In the first step is measured the similarity between clusters, using for instance the cosine or jaccard similarity. This \(d \times d\) matrix is then segmented into groups. We get clusters of clusters, or meta-clusters. In the final step, each item is assigned to the best meta-cluster.
This group is often called Graph partitioning. When we measure the overlap between clusters, there is no overlap between clusters belonging to the same partitioning. Therefore, we get a \(k\)-partite graph (if there are \(k\) base partitioning).
Illustration with a \(3\)-Partite Graph
“Clouds” represent clusters, i.e., a group of several items. Link’s strength depends on the overlap between clusters.
After node re-organization. Meta clusters appears.
Matrix representation. Grey color \(= 0\). There is no connection between clusters of the same partitioning.
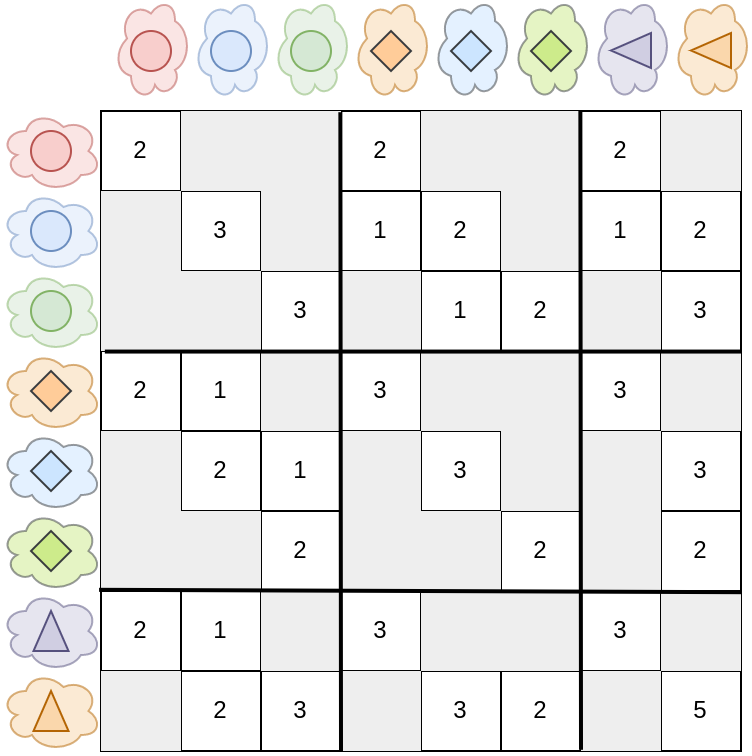
Note: Here, in this example, there are as many clusters as items. Therefore it looks as if both methods were equivalent. This is not the case, in practice \(n \gg d\)
Algorithms
For the algorithm, we have METIS, HGPA, CSPA TODO link
Hybrid
The hybrid form is the “rawest” one: in item-based approach, we forget about initial clusters, while in cluster-based, we forget about items. The hybrid version preserves almost all the information about the base clustering.
Each partitioning is represented as a binary matrix, using hot encoding.
\[H^{\ell}_{i, j} = \left\{ \begin{array}{ll} 1 & \mbox{ if } C^{\ell}(i) = j \\ 0 & \mbox{ otherwise} \end{array} \right.\]The final matrix \(H\) is the concatenation of all submatrix \(\{H^1, H^2, ..., H^k\}\).
Illustration
Here, gray blocks represent \(0\) and orange blocks \(1\).
Algorithms
Here, this representation is very similar to those of recommander systems, where co-clustering can be done.
Matrix factorization BCE Probabilistic
- Voting method
- Link based method (SimRank, Connected Tripple similarity)
Clustering ensemble based on sample’s stability
Evaluation of Ensemble Clustering
There are two ways to evaluate a clustering:
- external metrics, which compare base clusterings to the consensus clustering
- internal metrics, which relies on features / initial data \(X\) to evaluate the consensus clustering
In ensemble clustering, consensus quality is frequently evaluated using external measure. The reason is simple: EClust algorithms do not use feature \(X\) at any time, they only rely on the base clustering. It would be unfair to evaluate the partitioning on unavailable information.
External measure 1
These metrics are similarity measures, leading to a score between \(0\) (bad) and \(1\) (good):
- Normalized Mutual Information (NMI) 2
- Adjusted Rand Index (ARI)
The NMI is the most used in the literature, but ARI is still frequently seen. They have the advantage that they do not require the two partitioning to have the same number of clusters, nor to need to find a one-to-one matching between clusters.
In all these metrics, we measure the average score between the consensus clustering \(C^*\) and all base clustering \(C^i\).
\[Score(C^*) = \frac{1}{k} \sum_{i=1}^k Sim(C^*, C^i)\]Calinski-Harabasz Index Davies Bouldin
Internal measure
(For information, but useless in the context)
- Dunn Index
- Davies Bouldin
- Silhouette
- Compactness / SD index Halkidi
- Figure of Merit
- Stability
TODO: add them in clustering post
Diversity
TODO
Evaluation of ensemble is as easy as for clustering: we have metrics, we can apply them, however, this doesn’t means its truthful.
If you followed until them, this is the main proposition of this blogpost.
Usually, error metrics measure how much the consensus clustering lie in the middle of the base clustering.
\[\frac{1}{k} \sum_{i=1}^k (C^*, C_i)\]In classification, the poorest value you can have is \(\frac{1}{k}\) when choosing the class at random, if the dataset is balanced, and the best value is $1$, $100\%$ success.
The metric used for ensemble clustering are theoretically bounded by $0$ and $1$, so same as for accuracy and all others. Nevertheless, $1$ is not achievable. And the result (the average of the previous formula) depends on the number of clusters in each base clustering. And cherry on the cake, variation are of maximum $5 \%$.
I discover that by doing the following experiment: Instead of crafting a consensus clustering, use a base clustering to replace the consensus clustering. Then, the best base clustering is frequently the one with many clusters, and the difference of score between the two is very small.
So, what is a good clustering ?
Issues with external scoring method
General Experiment
The typical evaluation procedure of an ensemble clustering algorithm is the following:
- Pick a clustering algorithm (very often, \(k\)-Means)
- Create \(20\) base clustering. For each clustering, pick at random the number of cluster \(k\). Two options:
- \(k \in [n_{class}, \min(100, \sqrt{n})]\) (with \(n_{class}\) the number of classes in \(Y\), and \(n\) the number of items)
- \[k \in [n_{class}-2, n_{class}+2]\]
- Create the consensus clustering
- Perform evaluation.
Issues with the General Protocol
Evaluation Metrics
In classification, the best algorithm can get \(100\%\) accuracy. ARI and NMI are supposed to be scoring function between \([0, 1]\), which is the case. For consensus evaluation, the final score is the average similarity between the consensus and all base clustering. Here, it is impossible to get \(1\), unless your ensemble is made of the exact same partitioning (with \(0\) diversity).
Worst, when you pick one base clustering and consider it as the consensus clustering, the best obtained score is close to the algorithmic consensus result. You may swith from \(89\%\) to \(90\%\) (best base clustering VS consensus clustering). The difference between the two does not worth it for the computational effort.
Diversity
Here, diversity is obtained by changing the number of clusters. A simple reflection: if you create two partitioning, one with \(k\) clusters, the other with \(2 k\) clusters, It is possible that the \(k\) clusters are just splitted in two. In terms of diversity, that is not good (keep in mind that it is a possible option, there are some situation where this would not be the case).
Assuming this is the case (adding more clusters only split existing clusters), the results depend on the number of clusters.
We can look at NMI for that. NMI compare the entropy of two partitioning.
NMI
For \(k\) clusters of equal size:
\[H(C^k) = - \sum_{i=1}^k \frac{-\log_2{k}}{k} = \log_2(k)\]The conditional entropy for moving for \(k\) to \(s \times k\) cluster is:
\[H(C^k | C^{sk}) = - \sum_{i, j} P(C^k_i, C^{sk}_j) \log_2 P(C^k_i | C^{ks}_j) = \sum_{i, j} \frac{1}{sk} \log_2(s) = \log_2(s)\] \[H(C^{sk} | C^k) = - \sum_{i, j} P(C^k_i, C^{sk}_j) \log_2 P(C^{ks}_i | C^k_j) = \sum_{i, j} \frac{1}{sk} \log_2(1) = 0\] \[I(C^k, C^{sk}) = \sum P(C^k_i, C^{sk}_j) \log_2\frac{P(C^k_i, C^{sk}_j)}{P(C^k_i) P(C^{ks}_j)} = \sum \frac{1}{sk} \log_2\frac{\frac{1}{sk}}{\frac{1}{ksk}} = \sum \frac{1}{sk} \log_2(k) = \log_2(k)\]Therefore, the resulting NMI is:
\[NMI = \frac{I(C^k, C^{sk})}{H(C^k) + H(C^{sk})} = \frac{\log_2(k)}{\log_2(k) + \log_2(sk)} = \frac{1}{s+1}\]The NMI / ARI value mostly depends on the average number of clusters.
Appendices
Definition
- Hierarchical clustering
- Clustering algorithm performing \(n-1\) steps building the dependencies between items as a rooted tree
- Agglomerative clustering
- Kind of hierarchical clustering, where the initial step is made of \(n\) clusters with one item each and the final step has one single cluster with the \(n\) items. At each step, the closest clusters are merged together.
Bibliography
>> You can subscribe to my mailing list here for a monthly update. <<



